In a recent article we reported the performance of HADDOCK in the 2018 iteration of the Grand Challenge organised by the D3R consortium. Building on the findings of our participation in last year’s challenge, we significantly improved our pose prediction protocol which resulted in a mean RMSD for the top scoring pose of 3.04 and 2.67 Å for the cross-docking and self-docking experiments respectively, which corresponds to an overall success rate of 63% and 71% when considering the top1 and top5 models respectively.
This performance ranks HADDOCK as the 6th and 3rd best performing group (excluding multiple submissions from a same group) out of a total of 44 and 47 submissions respectively.
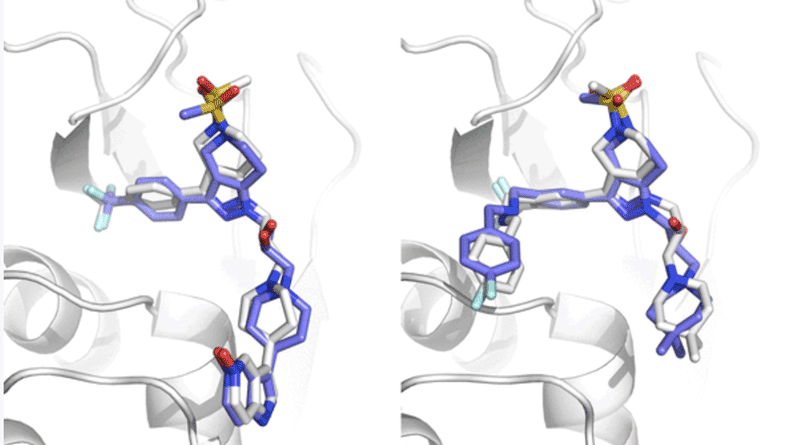
Superpositions of HADDOCK models on reference structures.
Left: model 5 from target 1 (1.1Å). Right: model 1 from target 8 (1.5Å).
Our ligand-based binding affinity predictor is the 3rd best predictor overall, behind only the two leading structure-based implementations, and the best ligand-based one with a Kendall’s Tau correlation of 0.36 for the Cathepsin challenge. It also performed well in the classification part of the Kinase challenges, with Matthews Correlation Coefficients of 0.49 (ranked 1st), 0.39 (ranked 4th) and 0.21 (ranked 4th) for the JAK2, vEGFR2 and p38a targets respectively.
Through our participation in last year’s competition we came to the conclusion that template selection is of critical importance for the successful outcome of the docking. This year we have made improvements in two additional areas of importance: ligand conformer selection and initial positioning, which have been key to our excellent pose prediction performance this year.
The data and code used to train the ligand-based binding affinity predictor and rank the compounds are freely available on GitHub, together with our in-house scripts developed during our participation in the last two GC competitions. These can be accessed at following URL: https://github.com/haddocking/D3R-tools.
Read all details in our publication:
- P.I. Koukos, L.C. Xue and A.M.J.J. Bonvin. Protein-ligand pose and affinity prediction. Lessons from D3R Grand Challenge 3. J. Comp. Aid. Mol. Des. Advanced Online Publication (2018).
